Peptide for Antibiotics and Nutraceuticals Design and Analysis
Funding programme: Bando Metadistretti /Biotecnologie, Regione Lombardia
Start – end year: 2006 – 2008
Partnership
ACS Dobfar S.p.A. (coordinator)
PRIMM S.p.A.
CELL Therapeutics Europe
University of Pavia (dip.to chimica farmaceutica)
University of Milano – Bicocca (dip.to bioscienze e biotecnologie)
Consorzio Milano Ricerche
FarmaOpera S.p.A.
The project
The project main objectives pursued along the research are to:
- Develop innovative biotech processes useful to produce molecules with wide application in different sectors like chemical, pharmaceutical and cosmetics.
- Optimise industrial biotech processes.
- Develop innovative computational methodologies appplied to the biotechologies.
The core competencies and activities of Milano Ricerche concern the set up of a structured system for data management, including information about toxicology, pharmacodynamics, pharmacocynetics and chemical of specific activity molecules.
The key factor of a structured system of data management is to aggregate and release research results in order to influence future investments both for research and development and production helping to better allocate those available resources in a profitable way.
A data framework developped in a multi-users perspective, provides also a fundamental tool for the condivision of results among many different interactive professional profiles. Using advanced technologies of machine learning and data mining allow to derivate information about molecules not yet experimentally measured. In this way, Knowledge Discovery attends the experiments design management.
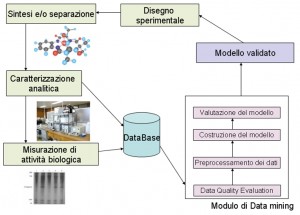
The following data integrated within the archive have an heterogeneous origin:
- Chemical data, coming from analytical instruments (peso molecolare, idrofobicità, solubilità, temperatura di fusione, coefficiente di ripartizione, strutture spettroscopiche, spettri di cromatografia HPLC),
- Pharmacodynamics data, coming from activities samples (risultati di saggi di binding molecolare o cellulare, EC50 in condizioni sperimentali),
- Pharmacocynetics data, related to ADMET parameters (biodisponibilità orale in organismi modello, dati di binding a proteine plasmatiche),
- Toxicology data (valori di legame ai recettori degli estrogeni, LC50 in specifiche condizioni, cancerogenicità in organismi modello),
- “in silico” data, coming from molecular modeling procedures.
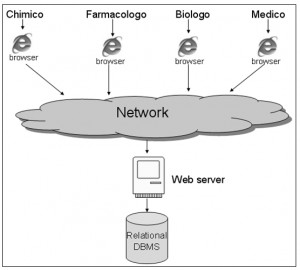
Finally, the overall system is realised through a multi-users web oriented architecture.
This is the general usage scheme of the archive.